Modeling dynamical biological processes through the lens of single-cell genomics
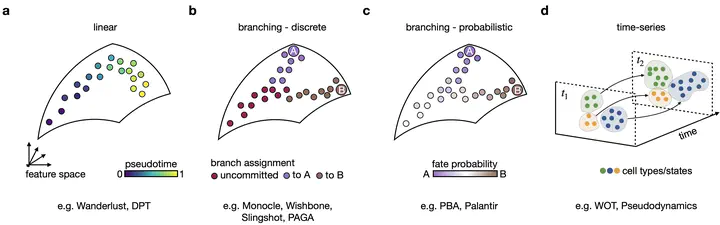 My thesis summarizes previous work on trajectory inference from single cell genomics data and introduces new methods that incorporate additional data modalities, including RNA velocity and lineage tracing information.
My thesis summarizes previous work on trajectory inference from single cell genomics data and introduces new methods that incorporate additional data modalities, including RNA velocity and lineage tracing information.Abstract
Cells dynamically change their molecular state in many situations. Single-cell assays allow us to describe this state with unprecedented resolution; however, cells are destroyed upon sequencing, making it difficult to study continuous processes. This difficulty has fueled the development of mathematical methods that use ensembles of cells to reconstruct latent trajectories. In this thesis, we present new approaches for trajectory reconstruction which build on recent experimental innovations.
Type
Publication
TUM online library