moscot preprint out
Mapping cells through time and space with moscot
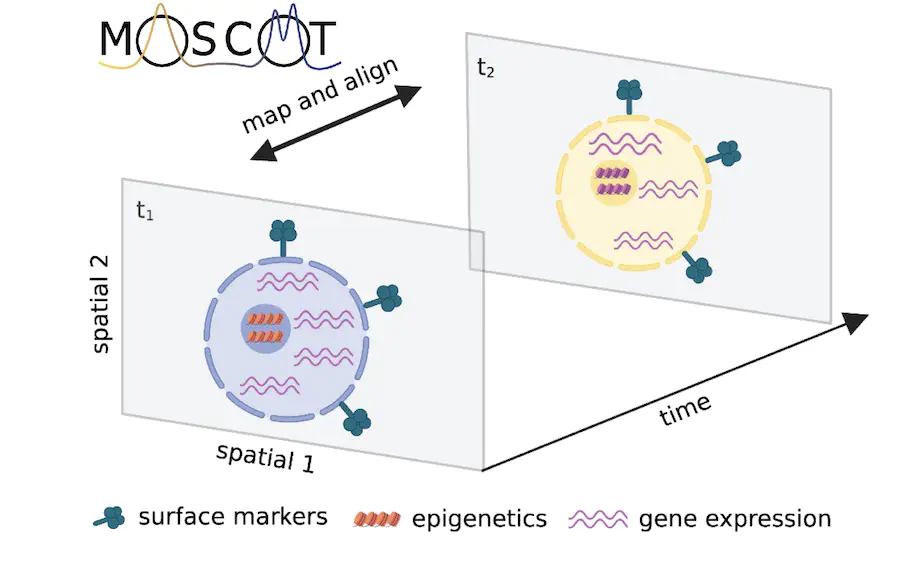
We’ve just released moscot, our new computational framework that maps single cells across time and space using efficient optimal transport algorithms. Importantly, moscot makes use of multi-modal information consistently across all applications, scales to atlases, and comes with a modular and extensible implementation. To find out more, see the tweetorials from Dominik or me, or read the full preprint on bioRxiv. Check out the implementation at moscot-tools.org to try it on your own single-cell or spatial data.
This project was co-lead by Dominik Klein, Giovanni Palla, and me from the Theislab, as well as Michal Klein from Apple ML Research and Zoe Piran from the Nitzan lab
Abstract
Single-cell genomics technologies enable multimodal profiling of millions of cells across temporal and spatial dimensions. Experimental limitations prevent the measurement of all-encompassing cellular states in their native temporal dynamics or spatial tissue niche. Optimal transport theory has emerged as a powerful tool to overcome such constraints, enabling the recovery of the original cellular context. However, most algorithmic implementations currently available have not kept up the pace with increasing dataset complexity, so that current methods are unable to incorporate multimodal information or scale to single-cell atlases. Here, we introduce multi-omics single-cell optimal transport (moscot), a general and scalable framework for optimal transport applications in single-cell genomics, supporting multimodality across all applications. We demonstrate moscot’s ability to efficiently reconstruct developmental trajectories of 1.7 million cells of mouse embryos across 20 time points and identify driver genes for first heart field formation. The moscot formulation can be used to transport cells across spatial dimensions as well: To demonstrate this, we enrich spatial transcriptomics datasets by mapping multimodal information from single-cell profiles in a mouse liver sample, and align multiple coronal sections of the mouse brain. We then present moscot.spatiotemporal, a new approach that leverages gene expression across spatial and temporal dimensions to uncover the spatiotemporal dynamics of mouse embryogenesis. Finally, we disentangle lineage relationships in a novel murine, time-resolved pancreas development dataset using paired measurements of gene expression and chromatin accessibility, finding evidence for a shared ancestry between delta and epsilon cells. Moscot is available as an easy-to-use, open-source python package with extensive documentation at https://moscot-tools.org.