CellRank 2 preprint out
Unified fate mapping in multiview single-cell data
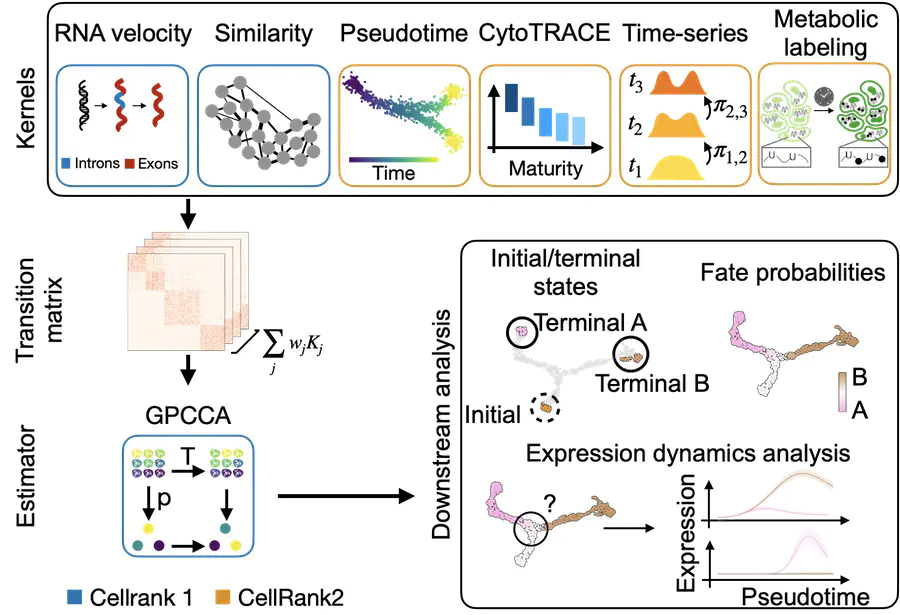
Happy to share that we released our preprint presenting CellRank 2, a unified framework to study cellular fate decisions. CellRank 2 comes with a modular interface that makes it easy to learn transition probabilities among cells based on various data modalities or views; currently, these include any Pseudotime, developmental potential, real-time information, and metabolic labeling data. In addition, CellRank 2 inhertis all of CellRank 1’s functionality to work with RNA velocity. CellRank 2 further accelerates version 1 and scales to millions of cells. Use it to compute initial and terminal states, fate probabilities, putative driver genes, gene expression trends along specific trajectories, and much more. Beyond the functionality we demonstrate in this preprint, CellRank 2 may also be applied to lineage-traced data downstream of moslin, or to spatio-temporal data downstream of moscot.
To find out more, see the tweetorials from Philipp or me, or read the full preprint on bioRxiv. Check out the implementation at cellrank.org to try it on your own single-cell or spatial data. CellRank 2 comes with a completely updated set of tutorials that make it easy to get started.
This project was co-lead by Philipp Weiler from the Theislab and me.
Abstract
Single-cell RNA sequencing allows us to model cellular state dynamics and fate decisions using expression similarity or RNA velocity to reconstruct state-change trajectories. However, trajectory inference does not incorporate valuable time point information or utilize additional modalities, while methods that address these different data views cannot be combined and do not scale. Here, we present CellRank 2, a versatile and scalable framework to study cellular fate using multiview single-cell data of up to millions of cells in a unified fashion. CellRank 2 consistently recovers terminal states and fate probabilities across data modalities in human hematopoiesis and mouse endodermal development. Our framework also allows combining transitions within and across experimental time points, a feature we use to recover genes promoting medullary thymic epithelial cell formation during pharyngeal endoderm development. Moreover, we enable estimating cell-specific transcription and degradation rates from metabolic labeling data, which we apply to an intestinal organoid system to delineate differentiation trajectories and pinpoint regulatory strategies.