Mapping lineage-traced single cells across time points (NeurIPS LMRL)
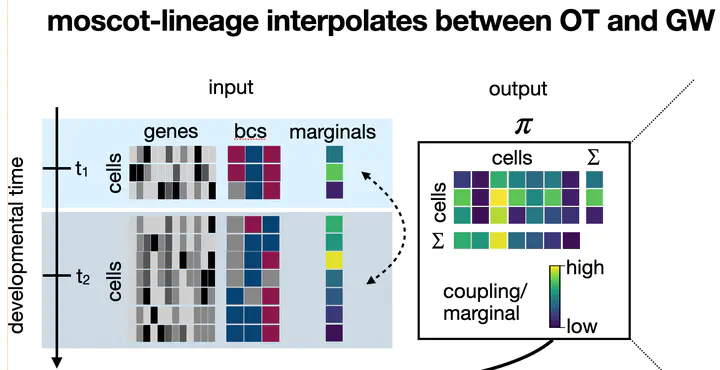 moscot-lineage maps lineage traced cells across time points using both gene expression and barcode information. Watch the recorded talk on YouTube
moscot-lineage maps lineage traced cells across time points using both gene expression and barcode information. Watch the recorded talk on YouTubeAbstract
Simultaneous profiling of single-cell gene expression and lineage history holds enormous potential for studying cellular decision making. However, it is currently unclear how lineage and gene expression information across experimental time-points can be combined in destructive in-vivo sequencing experiments. In this video, we present moscot-lineage, a Fused Gromov-Wasserstein based model to couple cellular profiles across time-points. In contrast to existing methods, moscot-lineage combines intra-individual lineage relations with inter-individual gene expression similarity. We demonstrate on simulated and real data that moscot-lineage outperforms LineageOT, the only competing approach. Further, on C. elegans embryonic development, we show how moscot-lineage, combined with existing trajectory inference methods, predicts fate probabilities and putative decision driver genes.
For more details, see our extended abstract.